|
TSSr: an R/Bioconductor
package for analysis of transcription start site
(TSS)
TSSr is designed to analyze transcription start sites (TSSs)
and
core promoters with most types of 5’end sequencing data, such
as
cap analysis of gene expression (CAGE) (Takahashi, Lassmann et al.
2012), no-amplification non-tagging CAGE libraries for Illumina
next-generation sequencers (nAnT-iCAGE) (Murata, Nishiyori-Sueki et al.
2014), a Super-Low Input Carrier-CAGE (SLIC-CAGE) (Cvetesic, Leitch et
al. 2018), NanoCAGE (Cumbie, Ivanchenko et al. 2015), TSS-seq (Malabat,
Feuerbach et al. 2015), transcript isoform sequencing (TIF-seq)
(Pelechano, Wei et al. 2013), transcript-leaders sequencing (TL-seq)
(Arribere and Gilbert 2013), precision nuclear run-on sequencing
(PRO-Cap) (Mahat, Kwak et al. 2016), and GRO-Cap/5’GRO-seq
(Kruesi, Core et al. 2013).
Source code and documentation is available on
GitHub: https://github.com/Linlab-slu/TSSr
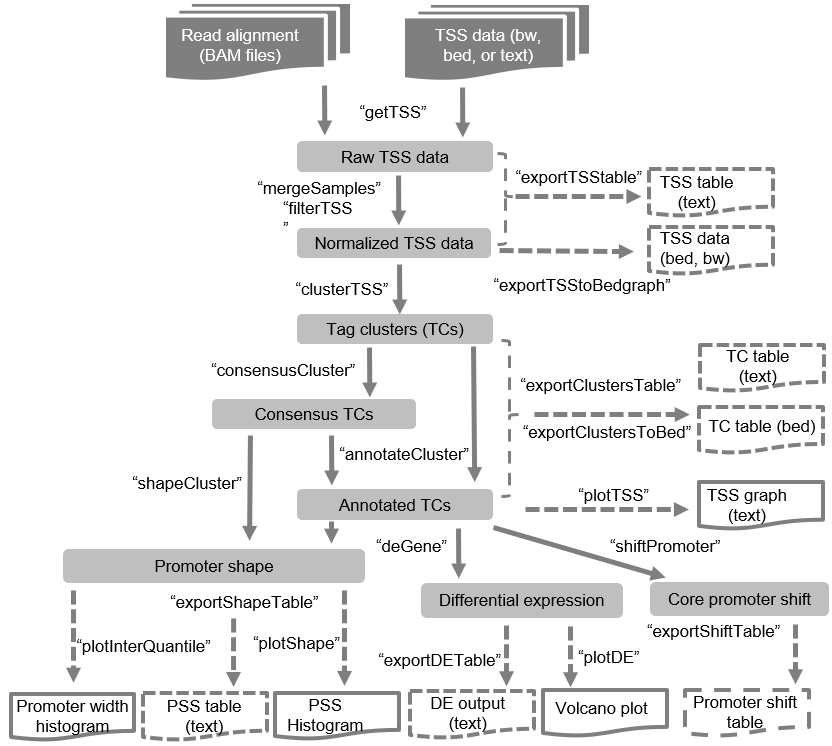
Overview of TSSr analysis
steps: importing primary raw data, processing data, clustering, and
integrative analyses.
Example data for TSSr
Example bam files
S01.sorted.bam
S02.sorted.bam
S03.sorted.bam
S04.sorted.bam
Example TSS table file
ALL.samples.TSS.raw.txt
Example genome annotation files
saccharomyces_cerevisiae_R64-2-1.gff
|





